By Michelle Wille, University of Sydney and Stacey Lynch, University of Melbourne
As we navigate a global human pandemic, avian influenza (or “bird flu”) has been detected in domestic poultry across Victoria.
When scientists discuss avian influenza, we’re usually referring to the diverse subtypes of influenza that primarily infect birds. Avian influenza viruses are commonly found in healthy wild birds and can also cause illness and death among domestic poultry including chickens, turkeys and ducks.
Humans can contract it if they come into close contact with infected birds (not from eating cooked chicken or eggs). But these viruses don’t easily infect us and their health risk is considered low.
Between 2003-2019, there have been about 2,500 human cases of avian influenza globally (mainly caused by the influenza subtypes H7N9 and H5N1).
There’s also no evidence of people becoming infected as a result of the current outbreaks in Victoria. Nonetheless, avian influenza viruses can mutate, so we must carefully monitor and deal with them as they arise.
Read more: 8 ways the coronavirus can affect your skin, from COVID toes, to rashes and hair loss
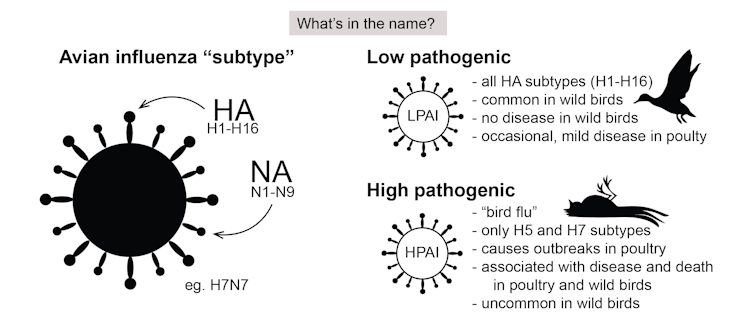
How we classify avian influenza
These viruses are classified in two ways. The first is based on the HA-NA subtype system. On the surface of the virus are two proteins: haemagglutinin (HA) and neuraminidase (NA). Of these, there are 16 and 9 types respectively.
So when we talk about the subtype H5N1, for example, we’re referring to type 5 of the HA and type 1 of the NA. Due to their mix-and-match nature, there are 144 potential HA-NA subtype combinations. The vast majority of these never cause disease in birds.
Avian influenza viruses are also classified by how “pathogenic” they are, which refers to their ability to cause disease in domestic poultry. Low pathogenic viruses are common in wild birds and may cause limited disease in poultry, but highly pathogenic viruses cause high mortality in poultry.
Occasionally, when an H5 or H7 low pathogenic avian influenza virus crosses from wild birds to poultry, changes in the virus genome can occur, transforming it into a highly pathogenic virus.
Avian influenza outbreaks in Victoria
In Victoria, there have been three outbreaks of avian influenza since July this year: two low pathogenic avian influenza viruses, H5N2 and H7N6, in domestic turkeys and emus, respectively, as well as a high pathogenic H7N7 virus in chickens.
The simultaneous detection of different virus subtypes in chickens, emus and turkeys is unusual. In the past, outbreaks in domestic birds have mostly been caused by a single subtype. This highlights the importance of stringent biosecurity practices, to prevent the introduction of avian influenza into farmed poultry.
Victoria’s current outbreaks are causing substantial economic loss and are considered emergency animal diseases. They have resulted in:
- the deaths of about 450,000 domestic birds across six farms, of which the vast majority are egg-laying chickens
- a potential loss of export markets for poultry products
- significant response costs and loss of income for affected producers, requiring permits to move eggs, equipment and birds from affected areas.
The good news is Australia has successfully eradicated high pathogenic avian influenza viruses in the past. We will almost certainly eradicate these too.
Agriculture Victoria, the lead agency for emergency animal diseases in the state, is responding to the outbreaks in a number of ways.
Firstly, a housing order requires all bird owners in the affected areas to keep their birds inside. This measure, along with other movement controls, helps limit spread to other farms.
Second, infected birds on the farms are destroyed, with the farms thoroughly decontaminated. These procedures are key to preventing the continued spread of avian influenza.

How we’re tracking the spread of the viruses
Outbreaks of avian influenza in Australian poultry are infrequent. The last outbreak of high pathogenic avian influenza in Victoria’s poultry (before this year) was in 1992. Low pathogenic avian influenza, however, is detected in our wild birds regularly.
Past testing has found different groups of wild birds can have infection rates ranging between 0.1-40%. The variation depends on which species make up the group, the group’s predominant location and also what season it is. The most common virus subtypes found in wild birds are H1, H3 and H6.
Data used to understand and monitor avian influenza in the wild is generated by the National Avian Influenza Wild Bird Surveillance program, which screens samples directly from captured birds, or indirectly through their faeces.
Not an ‘imported’ virus
Unlike contact tracing with people, birds can’t tell you who they have been socialising with. That’s why genomic sequencing is crucial in tracking, tracing and monitoring avian influenza viruses.
Each virus has a unique genomic sequence, like a genetic fingerprint. Using genetic analysis, the different genomes can be compared. This offers insight into how closely related certain viruses are and how wild birds may be spreading them across the country.
This method helped us to discover Victoria currently has three distinct outbreaks – and to connect the farms within each outbreak.
Also, a critical component of our response is the collection of virus genomes already available to us from past surveillance efforts. These data have revealed the viruses currently in Victoria are not imported from Asia, or elsewhere.
Rather, they’re similar to low pathogenic avian influenza viruses currently circulating in wild Australian waterbirds, as well as viruses that have caused past outbreaks in poultry.
We would like to acknowledge Agriculture Victoria, AgriBio, the Centre for AgriBioscience (a joint initiative between Agriculture Victoria and La Trobe University), the CSIRO Australian Centre for Disease Preparedness and the WHO Collaborating Centre for Reference and Research on Influenza in collaboration with Deakin University, for their ongoing avian influenza surveillance under the National Avian Influenza Wild Bird Surveillance program.
Michelle Wille, Australian Research Council Discovery Early Career Researcher Award Fellow, University of Sydney and Stacey Lynch, Senior Research Scientist, Agriculture Victoria, Honorary (Fellow) Veterinary Bioscience, University of Melbourne
This article is republished from The Conversation under a Creative Commons license. Read the original article.





